Abstract
Oncogenic signaling originating from the B-cell receptor (BCR) drives pathogenesis of most B-cell malignancies, including diffuse large B-cell lymphoma (DLBCL). KLHL6, encoding a substrate adaptor for Cullin-3 Ring-ligase (CRL) ubiquitination machinery, is involved in B-cell maturation and somatic mutations in KLHL6 have been reported in 5-15% of DLBCLs and other mature B-cell neoplasia. However, the function of KLHL6 and the consequences of its recurrent mutations in the context of B-cells and BCR signaling have been poorly established. Here, we have characterized the molecular basis of KLHL6 expression and mutations in DLBCL and applied high-throughput proteomic screening and functional characterization to determine the pathogenic role of KLHL6 dysregulation in DLBCL.
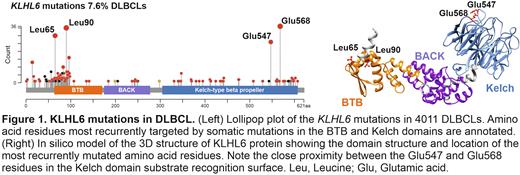
We observed that KLHL6 is localized on the perinuclear membranes in the reactive germinal center (GC) B-cells. This GC-like expression pattern was detected in 67% (n = 90/134) of the primary DLBCLs, and it was enriched in the GCB-cell-like lymphomas, of which 85% had the GC-like staining. By contrast, 44% of the activated B-cell-like (ABC) DLBCLs lacked this expression pattern, which associated with poor survival as compared to the ABC counterparts with intact GC-like KLHL6 expression. Loss of the GC-like expression pattern was associated with mutations in the MYD88 and other MCD/C5 driver genes, and mutations in the KLHL6 itself (p = 0.026). By pooled analysis of over 4,000 DLBCLs, we uncovered two classes of recurrent missense mutations that damaged either the N-terminal BTB- or the C-terminal Kelch-domain of the KLHL6 protein (Figure 1). On the protein level, loss of the GC-like KLHL6 expression pattern was restricted to BTB disruption, whereas Kelch-domain mutants retained physiological localization. Ectopic expression of the KLHL6 mutant proteins in the DLBCL cell lines confirmed these observations and revealed that BTB mutants abolish endogenous GC-like expression pattern in a dominant negative manner.
In malignant B-cell context, application of affinity-purification mass spectrometry (AP-MS) on KLHL6 baits revealed novel physical protein interaction partners outside the established ubiquitination/protein degradation machinery, such as BCR signaling components CD79A and Bank1. Moreover, we discovered that KLHL6 formed dimeric CRL-complexes with itself and two other BTB-Kelch family proteins, KLHL7 and KLHL26. These heterodimers were localized in separate compartments within B-cells and differentially induced upon BCR engagement. Notably, recurrent BTB-domain mutations disrupted heterodimerization but retained homodimerization, whereas Kelch-domain mutant KLHL6 retained all homo- and heterodimer conformations.
As a complementary approach to AP-MS, Kelch-domain adjacent proximity-dependent labeling (BioID2) revealed potential substrates processed by the identified KLHL6-CRLs. As an example, we observed that manipulation of KLHL6 expression altered the expression levels of CD79A in a substrate-like manner, translating to variable membrane expression levels of the BCR in DLBCL cells. BTB- and Kelch-domain mutant KLHL6 proteins retained different functional activities against the identified substrates. Notably, the recurrent BTB-mutants failed to promote CD79A degradation and, furthermore, the introduction of KLHL6L65P mutant increased the cell surface density of the BCR across DLBCL cell lines. By contrast, Kelch-domain hot spot mutants retained the ability to downregulate cell surface BCR levels. Further pathogenic properties of the KLHL6 mutants beyond surface BCR modulation were revealed by mutant protein proximity-dependent interactome screening. Overall, we discovered both shared and unique proteomic insults between the KLHL6 mutants that extended to processes not previously linked to dysregulation of BCR signaling in DLBCL.
In conclusion, we have discovered novel functional properties of KLHL6 in B-cells that are differently lost, retained, and endorsed by the recurrent mutants previously regarded solely as loss-of-function type. These results provide mechanistic understanding to the molecular pathogenesis of KLHL6 deregulated clonal B-cell outgrowths and establish a translational rationale for therapeutic interventions targeting BCR signaling in KLHL6 mutated B-cell neoplasia.
Disclosures
Leppa:Orion Pharma: Consultancy; Pfizer: Consultancy; Beigene: Consultancy; Bayer: Research Funding; BMS: Consultancy, Research Funding; Gilead Sciences: Consultancy, Honoraria; Roche: Consultancy, Research Funding; Novartis: Consultancy, Honoraria, Research Funding; Incyte: Consultancy, Honoraria; Genmab: Research Funding; Nordic Nanovector: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal